TWIST – Training With Input Selection and Testing (International Patent)
TWIST (Training With Input Selection and Testing) is a new evolutionary algorithm [1] able to generate two subsets of data with a very similar probability density of distribution and with the minimal number of effective variables for pattern recognition.
Consequently, in the TWIST algorithm every individual of the genetic population will be defined by two vectors of different lengths:
1) the first one, showing which records (N) have to be stored into the subset A and which ones have to be stored into the subset B;
2) the second one, showing which inputs (M) have to be used into the two subsets and which one have to be deleted.
The Figure depicts the dynamic of the TWIST algorithm.
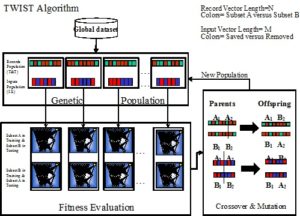
We have shown in many papers [2-15] that the TWIST algorithm outperforms the other splitting strategy (i.e. random distribution and K-Fold Cross Validation) in terms of results when they are applied to real medical data and also to classic datasets available from the UCI Machine Learning Repository [16].
The “reverse strategy” used in this algorithm tends to generate two subsets with the same probability density function, and this is exactly the gold standard of every random distribution criterion [3]. In addition, when the “reverse strategy” is applied, two fitness indicators are generated: the accuracy on the subset B after the training on the subset A, and the accuracy on the subset A after the training on the subset B. But only the lower accuracy of the two is saved as the best fitness of each individual of the genetic population rather than an average of the two or the higher of the two. This criterion increases the statistical probability that the two sub-samples are equally balanced during the genetic evolution because of the quasi logarithmic increase of the optimization process. We have also demonstrated experimentally [2] that when there is no information in a dataset, the behaviors of the TWIST algorithm, the Training and Testing Random Splitting and the K-Fold Cross Validation are absolutely equivalent. Therefore, TWIST does not code noise to reach optimistic results [2].
Our results have shown that TWIST algorithm is superior to current methods: pairs of subsets with similar probability density functions are generated, without coding noise, according to an optimal strategy that extracts the minimal number of features and the most useful information for pattern classification.
References:
[1] M. Buscema, “Genetic doping algorithm (GenD): theory and application”, Expert Systems, Vol. 21, No. 2, Blackwell Publishing, May 2004, pp 63-79.
[2] M Buscema, M Breda, W Lodwick, Training With Input Selection and Testing (TWIST) algorithm: a significant advance in pattern recognition performance of machine learning, in Journal of Intelligent Learning Systems and Applications, 2013, 5, 29-38.
[3] M. Buscema., E. Grossi, M. Intraligi, N. Garbagna, A. Andriulli and M. Breda, “An optimized experimental protocol based on neuro-evolutionary algorithms. Application to the classification of dyspeptic patients and to the prediction of the effectiveness of their treatment”, Artificial Intelligence in Medicine, Vol. 34, Issue 3, July 2005, pp. 279-305.
[4] S. Penco, E. Grossi et al., “Assessment of the Role of Genetic Polymorphism in Venous Thrombosis Through Artificial Neural Networks”, Annals of Human Genetics, Vol. 69, Issue 6, University College London, November 2005, pp. 693-706.
[5] E. Grossi, A. Mancini and M Buscema, “International experience on the use of artificial neural networks in gastroenterology”, Digestive and Liver Disease, Vol. 39, Issue 3, March 2007, pp. 278-285
[6] E. Grossi and M. Buscema, “Introduction to artificial neural networks”, European Journal of Gastroenterology &Hepatology, Vol. 19, 2007, pp.1046-1054.
[7] E. Grossi, R. Marmo, M. Intraligi and M. Buscema, “Artificial Neural Networks for Early Prediction of Mortality in Patients with Non-Variceal Upper GI Bleeding”, Medical Informatics Insights, Vol. 1, 2008, pp. 7-19.
[8] E. Lahner, M. Intraligi, M. Buscema, M. Centanni, L. Vannella, E. Grossi and B. Annibale, “Artificial neural networks in the recognition of the presence of thyroid disease in patients with atrophic body gastritis”, World J Gastroenterol, Vol. 14, No.4, January 2008, pp. 563-5688.
[9] S. Penco, M. Buscema, M.C. Patrosso, A. Marocchi and E. Grossi, “New application of intelligent agents in sporadic amyotrophic lateral sclerosis identifies unexpected specific genetic background”, BMC Bioinformatics, 9:254, May 2008.
[10] M.E. Street, E. Grossi, C. Volta, E. Faleschini and S. Bernasconi, “Placental determinants of fetal growth: identification of key factors in the insulin-like growth factor and cytokine systems using artificial neural networks”, BMC Pediatrics, June 2008, pp. 8-24..
[11] L. Buri, C. Hassan, G. Bersani, M. Anti, M. A. Bianco, L. Cipolletta, E. Di Giulio, G. Di Matteo, L. Familiari, L. Ficano, P. Loriga, S. Morini, V. Pietropaolo, A. Zambelli, E. Grossi, M. Intraligi, M. Buscema; SIED Appropriateness Working Group, “Appropriateness guidelines and predictive rules to select patients for upper endoscopy: a nationwide multicenter study”, American Journal of Gastroenterology, Vol. 105, No. 6, Jun 2010, pp. 1327-1337.
[12] M. Buscema, E. Grossi, M. Capriotti, C. Babiloni and P. M. Rossini, “The I.F.A.S.T. Model Allows the Prediction of Conversion to Alzheimer Disease in Patients with Mild Cognitive Impairment with High Degree of Accuracy, Current Alzheimer Research”, Current Alzheimer Research, Vol. 7, No. 2, Mar 2010, pp. 173-187.
[13] F. Pace, G. Riegler, A. de Leone, M. Pace, R. Cestari, P. Dominici and E. Grossi; EMERGE Study Group, “Is it possible to clinically differentiate erosive from nonerosive reflux disease patients? A study using an artificial neural networks-assisted algorithm”, European Journal of Gastroenterology & Hepatology, Vol. 22, Issue 10, October 2010, pp. 1163-1168.
[14] F. Coppedè, E. Grossi, F. Migheli and L. Migliore, “Polymorphisms in folate-metabolizing genes, chromosome damage, and risk of Down syndrome in Italian women: identification of key factors using artificial neural networks”, BMC Medical Genomics, 3:42, September 2010.
[15] G. Rotondano, L. Cipolletta and E. Grossi, “Artificial neural networks accurately predict mortality in patients with nonvariceal upper GI bleeding”, Gastrointestinal Endoscopy, Vol. 73, Issue 2, February 2011, pp. 218–226.
[16] A. Frank and A. Asuncion, UCI Machine Learning Repository, Irvine, CA: University of California, School of Information and Computer Science, 2010, http://archive.ics.uci.edu/ml.
Patents:
Training & Testing System : System and method for optimisation of a database for the training and testing of prediction algorithms. Applicant Semeion Inventor M. Buscema , USA Patent US 7,711,662 B2 – May 4 2010 (Application n. US60/440,210 deposited 01-15-2003). International Patent : Application n. PCT/EP04/00157 deposited 01-13-2004
